Background
Historically, acute myelogenous leukemia (AML) has been characterized as having a low somatic mutational burden and is predicted to have low neoantigen levels compared to solid tumors. However, recent data has shown that AML is an immunogenic malignancy and oncogenic driver mutations promote immune escape through immunoediting. Downregulation of human leukocyte antigen (HLA) class II HLA-DRB1, HLA-DQB1, HLA-DPA1 and HLA-DPB1 gene expression on leukemia cells has previously been associated with immune escape in AML following allogeneic hematopoietic stem cell transplantation (allo-HCT). While allo-HCT and cellular therapies are well-known potentially curative treatment options in AML, the human leukemia immunopeptidome has still not been well characterized. Here, we sought to uncover the human leukemia immunopeptidome by detecting differential major histocompatibility complex (MHC) class I neopeptide expression using molecular biotype subgroups.
Methods
We conducted retrospective analysis on 31 patients with hematopoietic malignancies including AML, chronic myelomonocytic leukemia (CMML), myelodysplastic syndrome/myeloproliferative neoplasm (MDS/MPN), MDS, chronic myelogenous leukemia (CML) and other hematopoietic malignancies (i.e. NK cell leukemia) at MedStar Georgetown University Hospital. Mutations were identified using next generation sequencing and separated into the following annotated molecular biotypes based on the mechanism of pathogenesis: Group 0 (G0) DNA repair, G1 splicing, G2 transcription factor, G3 kinase activation, G4 epigenetic modification, G5 chromatin remodeling, G6 cohesin, G7 tumor suppressor (i.e. p53), G8 telomere maintenance, G9 histone modification, G10 nucleotransport (i.e. NPM1) and G11 cell cycle regulation. FASTA nucleotide sequences were obtained by subjecting mutations to transcript expression and cDNA alignment in the National Center for Biotechnology Information (NCBI) gene database. MHC class I binding neopeptides were identified by mapping patient FASTA nucleotide sequences and HLA class I alleles in the Net MHC pan 4.1 database. Neopeptides were grouped by % Rank and categorized as weak binding (WB) if > 0.5 and strong binding (SB) if ≤0.5. Descriptive and inferential statistics were performed using SAS.
Results
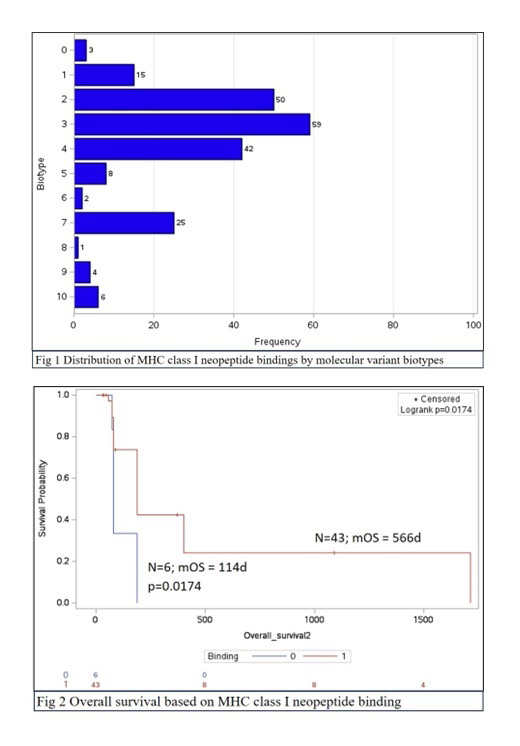
Thirty-one patients were evaluated including 17 AML (54.8%), 5 MDS (16.1%), 3 CMML (9.7%), 2 MDS/MPN (6.5%), 1 CML (3.2%) and 3 hematopoietic malignancies (9.7%). The median age was 65 years (range 28 -89); 43.8% were female and 43.3% were African American (AA). 144mutations were identified; thus far, 82 mutations have been subjected to FASTA NP_ protein extraction. The G3 biotype had the highest frequency of neopeptide bindings (n=59) followed by G2 (n=50) and G4 biotypes (n=42) ( Fig.1). Among patients with MDS, smokers were more likely to exhibit neopeptide binding compared to non-smokers (p=0.032). Patients with neopeptide binding had a longer median overall survival (mOS) compared to patients without neopeptide binding (mOS = 566 vs 114 days, p=0.0174) (Fig 2). Data exploration allowed isolation of a 54-year-old Asian male with Li-Fraumeni syndrome and p53 mutated AML who demonstrated high affinity neopeptide binding to HLA-B*39:01 with %Rank 0.08. Additionally, a 34 -year-old AA female with FLT3 mutated AML showed very high affinity neopeptide binding to HLA-A*29:02 with %Rank 0.007 as well as evidence of somatic HLA-A*03:01 deletion.
Conclusions
Pathogenic molecular variant biotypes constitute a novel immune signature, the human leukemia immunopeptidome. Smoking increased the likelihood of MHC class I neopeptide binding among patients with MDS. The presence of neopeptide binding predicted improved mOS in patients with myeloid malignancies. Further investigation of the human leukemia immunopeptidome may provide additional prognostic significance above that of standard molecular and cytogenetic risk stratification.
Disclosures
Renteria:Novo Nordisk: Consultancy; Kymera Therapeutics: Consultancy.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal